Single-Celled Organism Uses Internal ‘Computer’ to Walk
A Simple, Pond-Dwelling Protozoan Utilizes a Complex Walking Motion
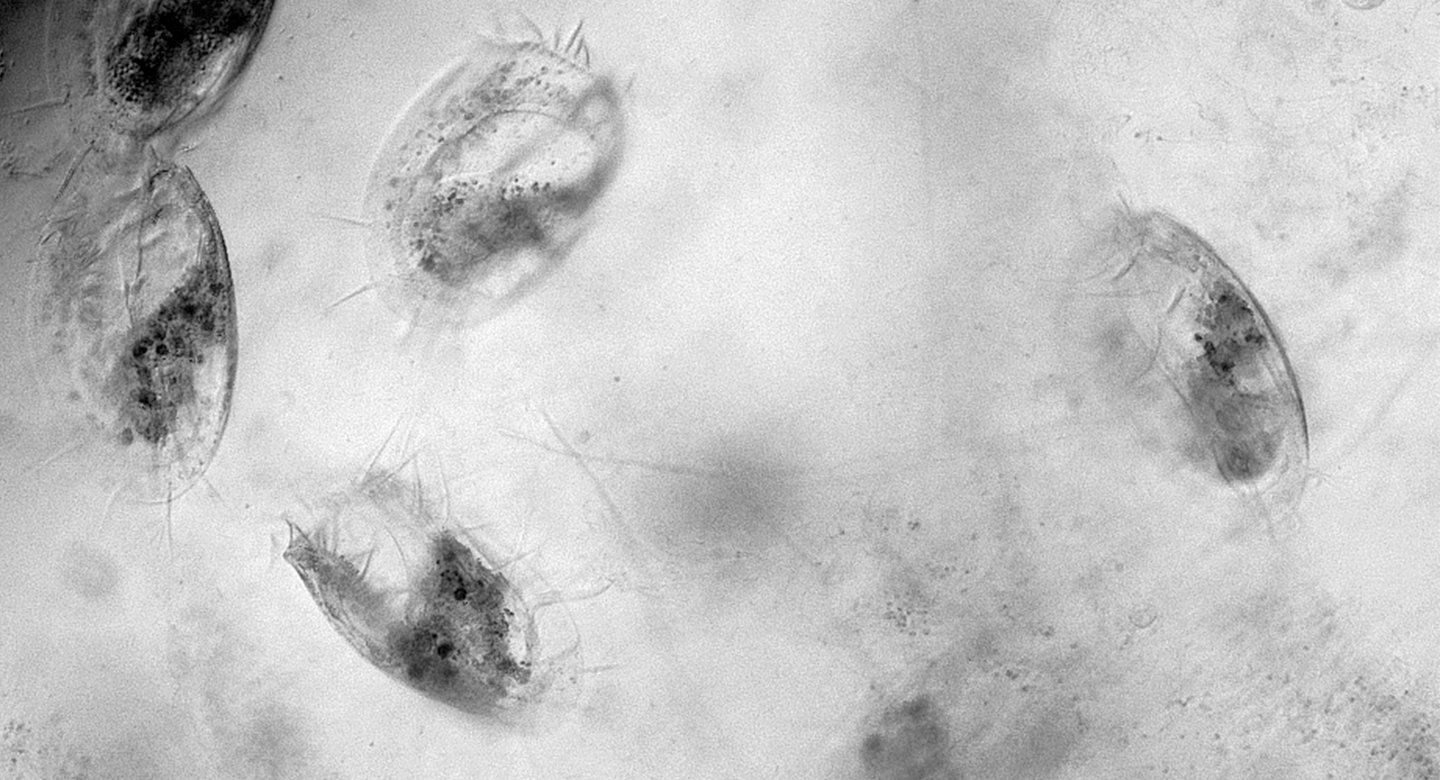
Most animals require brains to run, jump or hop. The single-celled protozoan Euplotes eurystomus, however, achieves a scurrying walk using a simple, mechanical computer to coordinate its microscopic legs, UC San Francisco researchers reported on September 22, 2022, in the journal Current Biology.
Euplotes has 14 leg-like appendages, each one composed of bundles of hair-like cilia. The researchers showed for the first time that internal connections between these cilia control their motions, letting the legs move only in certain patterns and sequences. When these internal connections are disrupted, the movements of the aquatic organisms become less productive – often leading the cells to turn in circles rather than walk in a line.
“Euplotes uses these connections to facilitate an elaborate walking motion, but my suspicion is when we delve into this more, we’ll find that other cells use similar forms of computation to control more subtle processes,” said Wallace Marshall, PhD, lead study author and professor of biochemistry and biophysics at UCSF.
In general, aquatic single-celled organisms don’t have legs and don’t walk – they roll, swim or slither. So when UCSF postdoctoral research fellow Ben Larson, PhD spotted Euplotes scurrying around under his microscope, he at first thought he was watching insect-like animals. When he realized the organisms were single-celled, he became intrigued by how they coordinated their 14 appendages without a brain or nervous system.
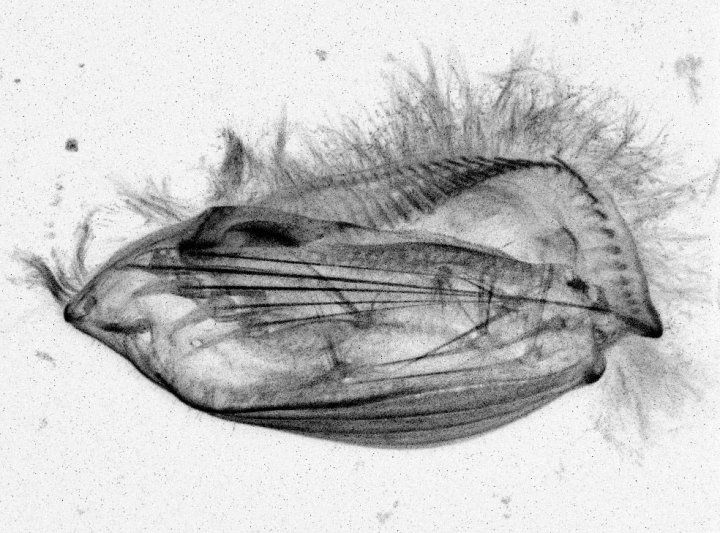
To understand this unusual ability, Larson and Marshall watched Euplotes cells in more detail, slowing down videos of the walking cells, capturing 33 frames per second, and labeling each leg to analyze the organisms’ gait.
The cells didn’t walk like people, with legs clearly alternating, nor did they have a cadence like a galloping horse. But Larson and Marshall found that the appendages did follow certain patterns. The researchers characterized 32 different “gait states,” or combinations of leg movements, and then showed that certain gait states were more likely to follow others.
“There seemed to be this sequential logic happening with the movements,” said Larson. “They weren’t random, and we began to suspect there was some sort of information processing happening.”
A Microtubule Machine
Scientists had known since the 1920s that long protein filaments projected into Euplotes from each of its appendages. Composed of microtubules – the main component of a cell’s scaffolding, or cytoskeleton – these filaments were long assumed to play a structural role in Euplotes. But when Larson and Marshall disrupted the microtubules with a drug or by cutting them with a needle, Euplotes no longer walked in the same way; its movements became more random and haphazard.
The researchers teamed up with computer scientists to model how the filaments could be controlling the walking motion. Together, they concluded that tension and strain on the filaments could dictate which gait states were possible at any given moment. The machinery, they said, resembles a Strandbeest – a moving, kinetic sculpture designed by a Dutch artist to walk and react to its environment.
Although this kind of internal machinery doesn’t resemble today’s digital devices, it does follow principles used by early mechanical computers, Marshall said.
“The fact that Euplotes’ appendages are moving from one state to another in a non-random way means this system is like a rudimentary computer,” he said.
More work is needed to understand exactly how the microtubule filaments control Euplotes walk, the researchers admit. But their computational modeling and experiments suggest a completely new, mechanical method for a cell to control its internal state.
“This is a really fascinating biological phenomenon itself, but also could highlight more general computational processes in other types of cells,” said Larson.
Co-authors: Jack Garbus and Jordan Pollack of Brandeis University were also authors of the paper.
Funding: The study was supported by an I2CELL Seed Award of the Fourmentin-Guilbert Scientific Foundation, the National Institutes of Health (R35 GM130327), the National Science Foundation (MCB- 2012647), and the Jane Coffin Childs Memorial Fund for Medical Research.