Building Blocks for COVID-19 Antiviral Drugs Identified in Rapid Study
Data Posted Online as an Open Resource for Scientists Developing Coronavirus Treatments
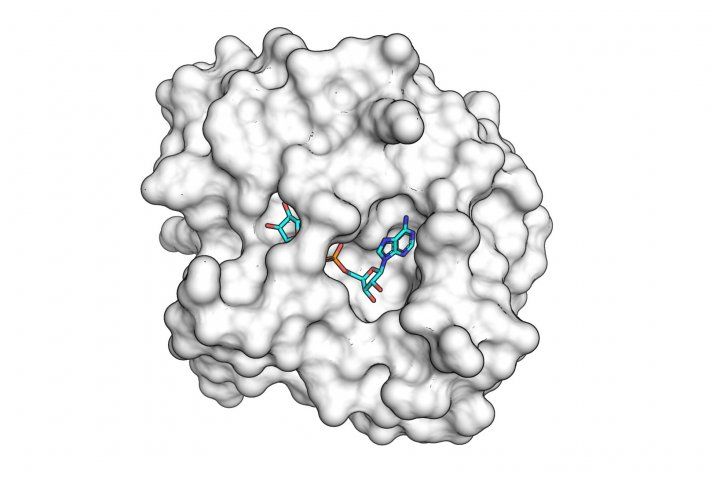
The SARS-CoV-2 macro domain protein bound to small molecule fragments that could be the basis of novel antiviral drugs.
Editor’s note: A manuscript detailing the findings was posted to the bioRxiv preprint server on Nov. 24, 2020, but has not yet been peer reviewed.
Scientists have identified key chemical building blocks for an eventual antiviral drug against SARS-CoV-2, the virus that causes COVID-19, according to data newly released by members of the UC San Francisco Quantitative Bioscience Institute Coronavirus Research Group (QCRG), based on research conducted with breakneck speed in collaboration with Lawrence Berkeley National Laboratory (Berkeley Lab) and SLAC National Accelerator Laboratory (SLAC).
The newly identified compounds bind to an enzyme produced by the virus, called the “macro domain,” which is known to be crucial for the virus’s ability to replicate in human cells.
“The SARS-CoV-2 macro domain is not as well understood as the virus’s main protease or the spike proteins that other efforts are going after,” said James Fraser, PhD, an associate professor in the Department of Bioengineering and Therapeutic Sciences, based in the UCSF schools of Pharmacy and Medicine, who led the research effort as part of the UCSF QCRG Structural Biology Consortium. “We wanted to look where others aren’t looking as heavily and have succeeded in identifying some promising candidates to build drugs that could halt the virus’s ability to replicate and spread in the human body.”
Further work is needed to weld these potential active ingredients into a workable drug candidate, but the research represents a promising new frontier in the battle against the virus, the researchers say, particularly given uncertainties about the timing or ultimate efficacy of an eventual vaccine.
The authors are writing up a formal manuscript describing the results for submission to a peer-reviewed academic journal, but also published their data directly online on July 1, 2020, to accelerate global efforts to fight the coronavirus pandemic. The open dataset was published in coordination with the Diamond Light Source X-Chem research group and Ivan Ahel’s lab at based at Oxford University, which simultaneously published their own parallel, complementary dataset of macrodomain-bound fragments.
“We’re delighted to openly share this data in coordination with the Diamond Light Source team, whose publication of a structure and fragment analysis of the SARS-CoV-2 main protease earlier this year was a huge step forward in galvanizing the computational chemistry community to develop additional antiviral targets,” Fraser said.
Novel Line of Attack on Coronavirus Biology
The study focused on an aspect of coronavirus biology that has only recently begun to be studied by researchers, including Alan Ashworth, PhD, FRS, president of the UCSF Helen Diller Family Comprehensive Cancer Center and a world leader in research on PARPs, enzymes that play a major role in DNA repair and have been targeted by drugs to treat certain cancers.
“PARPs can also reduce the effects of viral infection by tagging proteins with a molecule called ADP-ribose. As a countermeasure, some viruses – including the SARS family – have evolved macro domains that can remove this tag, re-exposing cells to infection,” Ashworth explained. “We are working to produce a drug to block the macro domain’s ability to remove these protective tags with the aim of reducing the impact of SARS-CoV-2 infection.”
Fraser and colleagues set out to understand the molecular structure of the macro domain in order to identify small molecules with the right shape to jam the viral protein’s active site, the first step toward building an antiviral drug that could inhibit the virus’s spread by re-empowering PARPs’ protective tags.
Using advances developed by the Fraser lab for a common but labor-intensive approach called X-ray crystallography, they team grew crystals of isolated macro domain proteins, then partnered with the Advanced Light Source at Berkeley Lab and the Stanford Synchrotron Radiation Lightsource at SLAC to bombard these crystals with powerful X-rays, using the resulting diffraction patterns to infer a detailed picture of the macro domain’s atomic structure down to a resolution of nearly one angstrom – one millionth the diameter of a human hair.
"The data quality was so incredible that, watching remotely, I initially thought the team was shooting a calibration sample," said James Holton, PhD, a faculty scientist in Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging Division who operates a beamline at the Advanced Light Source. "We almost never see resolution like that from the first ever crystal in a new project. It was truly amazing.”
“I am extremely impressed by the dedication of Fraser group in completing these vital experiments at record speed, added Aina Cohen, PhD, senior staff scientist at SLAC who co-leads SSRL’s Structural Molecular Biology (SMB) Division. “Their work demonstrates the importance of keeping essential synchrotron beamline facilities, such as those at the ALS and SSRL, operational during the recent shelter-in-place orders.”
“We were fortunate to partner with the two world-class light sources for X-ray crystallography located right here in the Bay Area,” Fraser said. “Using their amazing infrastructure, we were able to collect data from both light sources at once, which cut our research time in half.”
To identify small molecules capable of binding to the macro domain, the researchers used a technique called “fragment soaking” which involves creating hundreds of macro domain crystals, then using sound waves to manipulate candidate small molecules into the heart of each crystal and freezing them together in liquid nitrogen before returning to the X-ray light sources for further imaging.
X-ray diffraction experiments at Berkeley Lab and SLAC revealed 13 small molecules – each about a third to a quarter the size of a normal drug molecule – that successfully bound to the macro domain, including four that bound to the enzyme’s active site. “These candidates are very exciting,” Fraser said. “These are the first synthetic compounds to be identified that could make good starting points for a macro domain inhibitor drug capable of blocking viral replication in the human body.”
"What's so impressive about this is that crystallography is used here as a high-throughput approach," said Natalia Jura, PhD, an associate professor in the UCSF Cardiovascular Research Institute who leads the crystallography effort within the QCRG structure consortium with Fraser. "Typically, we use crystallography to solve a new structure based on a limited number of unique crystals that either will diffract well or not. If they fail, we are back to drawing board, optimizing crystallographic conditions and frequently data collection strategies. The infrastructure that’s been used here allows collecting hundreds of crystallographic datasets using well diffracting crystals that were soaked with drugs. It is an impressive data collection and data processing effort, only possible in recent years, and spearhead at UCSF by the Fraser lab.”
Using the new information about what chemical structures can bind effectively to the macro domain’s active site, Fraser and colleagues are now working with a virtual drug discovery platform developed by Brian Shoichet, PhD, a professor in UCSF School of Pharmacy’s Department of Pharmaceutical Chemistry, to rapidly screen through half a billion potential drug-like molecules to find drug candidates that combine several of these potent chemical structures.
“Fragment-based screening efficiently explores drug chemistry, using building-block ‘fragments’ from which drugs are synthesized,” Shoichet explained. “Using advanced x-ray crystallography techniques that the Fraser lab has pioneered, these studies provide a detailed map of how different drug components should interact with different parts of the viral macrodomain. Now, using a technology also pioneered at UCSF, we will computationally fit 500 million molecules onto these maps, seeking those that can stitch these different parts together in a single molecule, much increasing antiviral potency.”
Collaboration Reduced 2-Year Timeline to 10 Weeks
The study was conducted under the umbrella of the QCRG, which in April published a widely cited study that was among the first to map out the many ways in which SARS-CoV-2 interacts with the proteins it encounters when it infects a cell, which it followed up in June with a study showing how the virus hijacks key host proteins to promote its survival. The new study by Fraser and colleagues represents the first follow-up study from the QCRG’s structural biology working group aimed at identifying weak points in the virus’s infection mechanisms that could be targeted by drug development.
“It has been fantastic to the see the QBI community come together in the context of the QCRG,” said Nevan Krogan, PhD, the director of the UCSF Quantitative Biosciences Institute in the School of Pharmacy and a Gladstone Institutes Senior Investigator, who formed the QCRG in March. “It shows how fast we can move when we all band together and it especially exciting to see the younger trainees working together on such a pressing issue. The challenge will be to keep this spirit and infrastructure in place so that we can study all disease in this manner and hopefully be more prepared for the next pandemic.”
The project represents a remarkably speedy achievement for structural biology, according to Fraser, moving from cloning the gene for the viral macrodomain to completing the project in just about ten weeks.
“We were able pull this off on a hyper-aggressive timeline thanks to our many collaborators and especially to a grant from the UCSF Program in Breakthrough Biomedical Research – which let us stitch together a coordinated workflow between crystallography, the National Lab light sources, and UCSF Small Molecule Discovery Center – so the data could just flow in seamlessly,” Fraser said. “Without that philanthropic grant and the collaborative spirit of the QCRG, this would have been two-year project.”
Authors: See QCRG Structural Biology Group site for full list of contributors.
Funding: The research was funded in part by the UCSF Program in Breakthrough Biomedical Research (PBBR); the QBI Coronavirus Research Group (QCRG); a Sangvhi-Agarwal Innovation Award; a Byers Award for Basic Science; the William K. Bowes Biomedical Investigator Program; a Rapid grant from the US National Science Foundation (NSF); the US National Institutes of Health; and the US Department of Energy Office of Science, including through the National Virtual Biotechnology Laboratory, a consortium of DOE national laboratories focused on the response to COVID-19, with funding provided by the CARES Act.
Disclosures: The authors declare no competing financial interests
The University of California, San Francisco (UCSF) is exclusively focused on the health sciences and is dedicated to promoting health worldwide through advanced biomedical research, graduate-level education in the life sciences and health professions, and excellence in patient care. UCSF Health, which serves as UCSF’s primary academic medical center, includes top-ranked specialty hospitals and other clinical programs, and has affiliations throughout the Bay Area.