By Jeffrey Norris
Breast cancers differ from one another and from tumors that arise in other tissues. A drug that is life-saving for one cancer patient may fail in another with a tumor that originated in the same organ. But it's tough to match the patient to the best treatment.
"We don't have indicators for the vast majority of drugs to predict who should be getting them," says Joe Gray, PhD.
Gray, associate laboratory director for life and environmental sciences at the Lawrence Berkeley National Laboratory (LBNL) and co-leader of the breast cancer program for the National Institutes of Health-designated UCSF Comprehensive Cancer Center, is working with many collaborators to improve this state of affairs.
 |
Joe Gray |
Continued success in the collaborators' studies of cancer cells and human tumors will lead to smarter use of existing drugs. The research also should result in better informed development of new pharmaceuticals, and in the identification of potent cancer-fighting combinations of drugs.
Already, Gray's team has used molecular biology to re-categorize breast cancer in illuminating ways, and has identified subsets of breast tumors that are the most likely to recur soon after surgery - despite ongoing treatment.
 |
Dan Pinkel. Photo/Kaz Tsuruta |
Gray and colleagues discussed their latest results during several talks and a press conference at the annual meeting of the American Association for Cancer Research on April 14-17.
Elusiveness of "Magic Bullets" for Cancer
Historically, the goal of cancer pharmaceutical research, despite limited success, has almost always been to develop drugs that thwart a broad spectrum of cancers while sparing normal tissue.
This is no trivial problem. Genetic abnormalities that accumulate in successive generations of cells drive cancer. The mix of abnormally acting and abnormally produced proteins that arise from these genetic changes has much to do with whether tumor cells succumb to or resist treatment.
 |
Donna Albertson. Photo/Kaz Tsuruta |
Even when treatment is effective initially, a tumor's tendency to genetically mutate often enables it to engender cells that escape death and regrow.
Cells from different tumors often differ in the biochemical pathways they favor to help them survive, grow and spread. And cancers can change course biochemically in an evasive response to treatment, with different tumors adapting in different ways.
As a result, the goal of developing a single drug that acts as a roadblock on an indispensable biochemical pathway used by all cells in all tumors has long remained elusive.
New Approach to Cancer Drug Development Evolves
In general, new drugs for cancer that are tested in broad-based clinical trials have continued to provide little overall survival benefit. Even so, many of these drugs substantially improve outcomes for a minority of patients.
 |
Frederic Waldman |
Pharmaceutical company leaders have started to rethink their approach, with an aim to develop drugs designed to more effectively target tumors with specific characteristics.
Many biochemical pathways that play a role in cancer are governed in part by proteins that already have been identified. And many of these biochemical outlaws have been targeted for drug development.
For instance, extra copies of the gene called HER2 are abnormally present in the cells of between 20 percent and 30 percent of breast tumors. HER2 is targeted by a drug called Herceptin, made by Genentech. A decade ago Gray and UCSF colleague Daniel Pinkel, PhD, developed a quick lab test to determine whether a tumor has amplified HER2. Herceptin normally is used only when tumors test positive for HER2.
Microarrays Reach the Mainstream
In more recent years, researchers have been developing the capability to obtain information simultaneously on many different genes or gene products obtained from cells.
One goal is to identify patterns of gene activation or inactivation that help predict how aggressive or deadly tumors are likely to be, and to ratchet up post-surgical therapy accordingly.
Gene activity in fresh tumor tissue now can be measured with a microarray "chip," essentially many thousands of lab tests on a glass slide.
 |
UCSF researcher Koei Chin, PhD, is a leading participant in a large collaborative project involving several laboratories and research institutions - research aimed at identifying differences in molecular profiles and drug responses among different cancer cell populations. The findings are leading to ways of matching individual cancer patients with treatments that are more likely to produce good results. Photo/Mark Serr
|
The MammaPrint - a microarray developed through cancer genetics research conducted by Laura van't Veer and colleagues at the Netherlands Cancer Institute (see
UCSF Cancer Center Report vol. 5.2, p. 8, Fall 2003) - became the first microarray to be approved for sale in the US as a clinical lab test earlier this year. MammaPrint, which measures activity of 70 selected genes, is used to help gauge cancer severity in women diagnosed with small breast tumors that have not yet spread to lymph nodes.
But even for drugs that have been available for many years, oncologists generally have no tests to signal which treatments are most likely to improve outcomes for specific patients with cancer.
There are about 100 FDA approved anti-cancer drugs, Gray notes. "Most of them have not been carefully tested in breast cancer," he says, and they certainly have not been tested for effectiveness against breast cancer subsets, he adds.
New Prognostic Indicators, New Cancer Drug Targets Identified
Using techniques developed at UCSF by Donna Albertson, PhD, Pinkel and colleagues, UCSF and LBNL investigators no longer needed to limit their searches for cancer genes - by focusing only on genes involved in cell division, for instance. Instead, the team reclassified breast cancers, based on comprehensive analysis of gene copy-number and gene activation. They identified distinct categories of breast cancers associated with a poor response to current therapies.
The LBNL and UCSF team then assembled a collection of approximately 50 different breast cancer cell cultures that have a wide range of molecular features, some associated with drug sensitivity and some with resistance. They now have used these cancer cell lines to develop molecular signatures associated with response to drugs such as Herceptin and Tykerb, a drug made by GlaxoSmithKline (GSK).
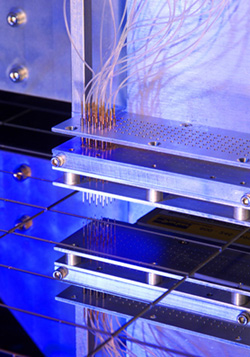 |
Microarray analysis locates genetic abnormalities in cancer cell chromosomes. An automated microarray spotter places DNA onto glass slides. Each spot contains DNA from a different chromosomal region. Photo/Mark Serr
|
In addition, by working with the cancer cells growing in the lab, and by validating their findings with data on real breast tumors, a group led by Gray and UCSF professor of laboratory medicine Frederic Waldman, MD, PhD, also identified dozens of genes in the chromosomes of many breast cancers - genes that not only are frequently amplified but that also appear to contribute to a poor prognosis. Based on predictions about the structures and functions of the proteins encoded by these genes, Gray's group identified nine that may be promising new targets for drug development.
Helping Big Pharma Make Better Picks
"Some of the targets we identified in breast cancers are in pathways well-known to play a role in cancer, but the roles of others were completely unknown," Gray says.
Long before GPS, explorers' maps included
terra incognita, with drawings or annotations to indicate "here be monsters." It's still early days in this renaissance of cancer gene-mapping. The roles of many monstrously-acting proteins that contribute to the many types of human cancers - and which may be potential targets for drug development - remain to be discovered.
But the pace of exploration has quickened exponentially, and better matching of treatment to cancer need not await an exhaustive accounting.
In the next step, a new research collaboration funded by the National Cancer Institute and in part by GSK, Gray and his university colleagues will be testing scores of drugs against the breast cancer cell lines to help guide which drug candidates will be selected for clinical trials. They will confirm the validity of the molecular markers associated with drug response in clinical samples.
The research collaboration should also lead to new strategies for combining drugs. Even as these comprehensive studies begin to shape new drug development and help to identify more effective uses for existing drugs, Gray says, "We know in the end that we are going to have to use combinations of drugs to be fully effective in treating many cancer patients."
Related Links:
Avon Project Symposium - Genetic Promise and Practice (PDF)
UCSF Cancer Center Report (vol. 5.2, p. 8), Fall 2003
A Collection of Breast Cancer Cell Lines for the Study of Functionally Distinct Cancer Subtypes
Cancer Cell 2006;10(6):515-527
Summary |
Full Text |
Full Text (PDF)
Genomic and Transcriptional Aberrations Linked to Breast Cancer Pathophysiologies
Cancer Cell 2006;10(6):529-541
Summary |
Full Text |
Full Text (PDF)
Breast tumor copy number aberration phenotypes and genomic instability
BMC Cancer 2006; 6:96
Abstract |
Full Text |
Full Text (PDF)
Breast Oncology
UCSF Comprehensive Cancer Center
Gray Lab
Ernest Orlando Lawrence Berkeley National Laboratories





