Poring Over Proteins: A Conversation with Protein Expert Andrej Sali
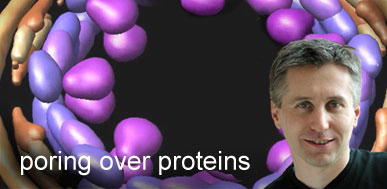 What does the movie illustrate? Localization of major substructures and their component nups in the nuclear pore complex. The nups are represented by their localization volumes and have been colored according to their classification into five distinct substructures based on their location and functional properties: the outer rings in yellow, the inner rings in purple, the membrane rings in brown, the linker nups in blue and pink, and the FG nups (for which only the structured domains are shown) in green. The pore membrane is shown in gray.
What does the movie illustrate? Localization of major substructures and their component nups in the nuclear pore complex. The nups are represented by their localization volumes and have been colored according to their classification into five distinct substructures based on their location and functional properties: the outer rings in yellow, the inner rings in purple, the membrane rings in brown, the linker nups in blue and pink, and the FG nups (for which only the structured domains are shown) in green. The pore membrane is shown in gray.
Good science takes time. And even after years of work, success is never guaranteed. That’s why there is both relief and joy in the lab of Slovenia-born Andrej Sali, PhD, a professor in the School of Pharmacy’s Department of Biopharmaceutical Sciences.
In November 2007, Sali co-authored two papers in the journal Nature. While the titles are a bit esoteric – “Determining the Nature of Macromolecular Assemblies” and “Molecular Architecture of the Nuclear Pore Complex” – their implications are rock-star huge. In short, after eight years, Sali and his colleagues have created software that takes information gathered from different sources, such as nuclear magnetic resonance spectrometry, X-ray crystallography and electron microscopy, to calculate the structure of protein complexes.

Why is this important? Well, in the case of the one big protein assembly known as the nuclear pore complex, Sali’s software – called IMP for integrated modeling platform – determined the positions of all 456 proteins, not just their overall shapes, in real time.
Keep in mind that proteins are fluid and dynamic, moving in a combined quadrille and quickstep. Seeing all the moves as they happen – which is Sali’s accomplishment – is essential to understanding the underlying choreography.
In this grand ballet, the nuclear pore complex is the first dancer; it is the gateway in and out of the cell. Knowing how to open and close that gate is a key piece of information for potential drug makers. And since function follows form, Sali’s work has laid bare the details.
It is precisely the kind of computational wizardry that Sali came to UCSF to perform. And in finding like-minded colleagues in the California Institute for Quantitative Biosciences, otherwise known as QB3, as well as in the School of Pharmacy, Sali has added new intellectual weight to an already critical mass.
Photo/Majed
Alternative content
Related Links
- Sali and Colleagues Create Technique to Reveal Architectures of Large Protein Assemblies
- Andrej Sali Lab
- Determining the architectures of macromolecular assemblies (PDF)
- Nature, November 2007
- The molecular architecture of the nuclear pore complex (PDF)
- Nature, November 2007
Podcast transcript
Miller: I’m Jeff Miller, welcome to Science Café. Today I’m with Andrej Sali, professor and vice chair of the department of Biopharmaceutical Sciences in the UCSF School of Pharmacy, and a faculty affiliate of the California Institute of Biosciences, otherwise known as QB3 here at Mission Bay. Welcome Andrej.
Sali: Thank you.
Miller: Well, when I read department of Biopharmaceutical Sciences or faculty affiliate of QB3, I don’t really know exactly what that means, so could you tell me what is it you do everyday?
Sali: Well, you come in the morning and you have a day that consists mostly of meetings with other scientists, primarily those in your own research group, students and postdocs, and you try to give advice and you try to learn from them—
Miller: What do you like best about your work?
Sali: I actually think it’s the very meetings I described. It’s talking about the scientific projects you’re involved in with the people who are executing them; planning for this research, and writing it up in scientific papers which are hopefully then published, I think it’s that process of planning and doing science that is most attractive — which is why most of the scientists are in science.
There are unfortunately a lot of other things that are necessary, such as raising money to fund all of this that by itself is not really pleasurable, at least not for me, but is kind of necessary.
Miller: I know you’re a protein guy, right?
Sali: That’s correct.
Miller: So why UCSF, and why, among all the many things you could have chosen to study did you choose to come here and then to study protein structure as well?
Sali: I’ll take the second part first, ‘why proteins’: I’m not sure I really know the answer why — I know that I was interested in proteins from high school on, I worked in a local lab in Slovenia, I’m originally from Slovenia. As a high school student I headed my own project which had to do with proteins in lab as well as on the computer, and it was always proteins — they’re just such an amazing combination of complexity and simplicity..
Miller: I have to ask you, is there anyone in your family who was a scientist?
Sali: My father has been a professor of psychology at the local university.
Miller: He didn’t influence your love of proteins?
Sali: I don’t think so. There was a biology textbook, if I think even further back, that we used in high school which was based on an American program in biology — I remember the introduction of the textbook used to educate Americans in the sciences, and one result of that was a great series of textbooks, and we used them.
This was Yugoslavia then, and it was just the most amazing textbook. And perhaps the love of proteins comes from that.
Miller: We’re going to talking a lot about proteins with you here today, but at the beginning, if we can get the numbers straight — how many proteins are there in the human body — do we know?
Sali: We know approximately that the human genome codes for about twenty-five thousand or so proteins, but many of them are then modified after they’re expressed, and so you have a number of different versions of the same protein encoded by the same gene, so the number is in excess of twenty-five thousand.But it’s also true that at any given moment in time, in any given cell, only a subset of them are really expressed. So perhaps you have only an order of 10,000 of so proteins that will exist and act at any given moment in any given cell.
Miller: You mentioned the human genome, I remember when the number of genes first came out for humans, approximately 30,000, some were surprised that it was so low. But isn’t it true that it’s the protein interactions that really allow for all this sophisticated development within humans?
Sali: Absolutely, I think that’s exactly right. The complexity, and you can see in biology or in evolution—if you study evolution—how complexity arises not so much from the number of genes or proteins that an organism encodes and uses, but from the interactions between these proteins, and also actually from the interactions between proteins and other molecules such as nucleic acids as well as various small molecules, be it substrates, or regulators and inhibitors of the activities.
So definitely a lot of complexity arises from the interactions between the components, not from the sheer number of components only.
Miller: So do we have any idea about how proteins actually evolved — it might not be a fair question, it might not be your field, but I’m always curious?
Sali: No no, it is a very relevant question, and one can describe many aspects of protein evolution from comparison of proteins from different organisms. Present day organisms, and you can in fact infer to a significant degree, at least in some situations, the exact changes, or almost the exact changes in nucleic acid sequences that resulted in the present day versions of proteins from the ancestors in the distant past, maybe hundreds of millions of years ago.
In fact a lot can be learned from comparison of, say, the human genome and the mouse genome, the human genome and primate genome. It’s a very fruitful area of biology that’s very informative about not just how we came to be what we are, but also about how the present day proteins function.
Miller: Do we know what the first protein was?
Sali: I don’t think so-
Miller: The mother or the father protein, we don’t have that?
Sali: There was almost certainly many more than one, they were probably small, single domain, very stable proteins, but exactly what they were, I’m not sure we’ll ever know.
Miller: Can we guess that maybe that the function of proteins has something to do with the energy needs of living organisms, or is that not related at all?
Sali: That is definitely one of the major functions, involved in generating the energy or the molecules that store the energy that is then used in other processes in the organism also regulated by proteins, but it’s just one of their functions.
Other major functions include, even just the structural framework for the organism of the cell — whole organism — as well as regulation of all sorts of processes, such as conversion of genes into proteins ultimately; There are inhibitor proteins that inhibit processes; There are enzymes that catalyze processes; There’s a very large variety of different kinds of functions and reactions that proteins are involved in.
Miller: I know proteins were the center of two November papers in the journal Nature, let me read the titles here, one was “Determining the Nature of Macromolecular Assemblies” and “Molecular Architecture of the Nuclear Pore Complex”, and then there was a Nature review in December, “The Molecular Sociology of the Cell”, but before we get into detail about those, I just want to spend a second to secure in people’s minds the dynamism of the cell.
I think sometimes the public, because they see static representations of cells and cell activity and cell proteins, they think of this as a fixed and static process when in fact it’s quite a dynamic one, if I’m reading this all correctly... And that there’s this constant communication and interaction. That’s really the hard thing to disentangle. And that’s really one of the aims of your research, correct?
Sali: Yes, I think that you’re absolutely right; proteins are dynamic entities, all sorts of levels and all sorts of different time scales. You have individual atoms, fluctuating around their average, or equilibrium positions, then you have larger parts of proteins moving with same structural elements, for example moving with respect to each other in one protein molecule. And then you have, of course, sometimes, the whole protein unfolding, for example when it’s degraded by some other protein protease, in that case, or you can also have whole protein molecules coming together and falling apart in larger complexes. So you have all those dynamic processes, many of which are important for their functions; some may be coincidental, but many of them are important for function and certainly we would like to have computational models for describing all these dynamics and models that allow us to regulate the dynamics as well as exploit it.
Miller: So for those that don’t know, could you explain what a computational model is?
Sali: There are different kinds of computational models, some of them will allow you to incorporate information from experimental sources, experimental methods that people generate in a lab. As they incorporate that information they will also generate some answers that are not directly observable in the lab about the system you’re interested in, about the protein you’re interested in. For example, in structures that can be determined that way, you measure certain aspects of the structure in crystallography that will be structure factors, perhaps phases, or estimated phases, and then you would get out of that an indirect set of observations, the actual three-dimensional structure of the protein. You need a computational model to allow you to convert the data into the structure.
That’s one kind. Another kind of model (and this classification is totally arbitrary I suppose) is totally self-contained, where you actually don’t rely, at least not directly, on any measurements about the system you’re interested in, but you have enough physics and perhaps enough statistical inferences — you incorporate it into the model, so that you can do everything that you want to do — predict the structure, as opposed to determine it, or simulate the motions as opposed to measure them. You do this entirely on computer.
Miller: And this is all about being able to “see” the proteins—
Sali: Yes exactly. One can obtain a very explicit microscopic picture of protein structures as well as how they change in time with such methods. It’s another question how accurate these pictures are, but with time they’re getting more accurate and hopefully…
Miller: -It’s seeing them in real time — these are moving and changing and interacting all the time...
Sali: That’s exactly right, yes.
Miller: And the, seeing them tells you what?
Sali: It depends on each case I suppose.
Miller: And when we talk about seeing them, the data can be interpreted to computer images?
Sali: That’s usually the case. In the old days when computer graphics was not powerful enough, people would build physical models of protein structures from plastics, but that’s not really done anymore, so it’s all on the computer screen. And there’s very powerful software that exists for such tasks.
Actually, one particular program that we like to use is called Chimera and it’s being developed by Tom Ferrin’s group here at UCSF. Many other people use it too. So it’s possible to do that on a computer screen, and it is an amazing property of structural biology that, in some sense, it’s justified to have almost a belief, a blind belief that when you see a protein structure, for the first time, you will have an insight, or a set of insights into how it acts, how it evolved. How you might interfere with it that you can’t even imagine, before you know what the structure is…
Miller: Is that true?
Sali: In many cases it’s true, it’s not always true.
Miller: So you just see it and something clicks.
Sali: That’s exactly right.
Miller: Based on your knowledge of other proteins perhaps?
Sali: That could be involved… It’s very difficult to be algorithmic about it, or to have a specific set of rules exactly how to proceed, but there’s a variety of possible insights and by seeing the structure one frequently has them, it’s just an empirical fact. One of the very beautiful features of structural biology is that function does depend on the form, and if you know the form you’re in good shape to get an insight into function.
Miller: OK, as long as we’re talking about form, let’s just say I’m sitting next to you on a plane and I realize that you’re one of the co-authors of this paper on the nuclear pore complex, how are you going to—
Sali: Not likely to happen (laughter)
Miller: Let’s just say, hypothetically speaking — so what are you going to tell me that is going to make me get excited about knowing the structure of the nuclear pore complex?
Sali: A number of things. You see the structure existing in the shape of a ring, with a hole in the middle, and that ring in fact sits in the nuclear envelope which defines the boundaries of the nucleus. And all of — because you know that most of the traffic — micro molecular traffic in and out of the nucleus goes through the nuclear pore complex, you can begin studying that hole in the middle of the ring in terms of the mechanism of transport.
Miller: Things get in and go out—
Sali: Through this gate of the nucleus, so called. And because you now have the structure, you can understand that there is transport factors involved in carrying the cargo molecules in and out through the ring you can begin to model the process quantitatively. Again in the computer. Hopefully informed by experiments that determine interactions between at least some of the components of the system.
But basically you are in a position to start describing quantitatively and then perhaps regulating the transport of different macromolecules differentially through the pore. In and out of the nucleus. And because it’s obviously very key to the function of the cell, what’s in and out of the nucleus, you can imagine all sorts of basic biology as well as biomedical applications.
Miller: Before we get into the potential biomedical applications, I just want to make sure you get credit for your role in this process, in this study, which if I understand it correctly, there are a lot of clues from these various technologies that infer or demonstrate what the potential shape is. Your role was to actually design software that was able to integrate all this information and then to come up with a proposed shape and eliminate a lot of other possibilities? Is that why people are so excited?
Sali: I would say that’s one of the reasons — well I hope that’s one of the reasons why people are excited… But it is absolutely true that the structure that was determined was determined based on experimental information about the nuclear pore complex. And that experimental information was of many different types, and was generated over the course of many years or so by our collaborators in Rockefeller University, New York City and students and post docs in their labs. The information it generated was very difficult to obtain and they generated a lot of it, and should be another reason why people should be excited about this whole thing.
So when you put the two together, the way to generate this kind of information and the software that allows you to convert information into an explicit three-dimensional structure, then you have a whole approach that should be, hopefully, applicable to many other complexes.
Miller: And you were the software conversion group —
Sali: And we developed the method and implemented it into software and applied it with our friends to this specific system of nuclear pore complex.
Miller: You mentioned earlier about potential biomedical applications, so what might some of this lead to? And you can speculate wildly, that’s OK.
Sali: Of course I have to preface what I’m saying, that it takes more than ten years on the average to develop a drug even when you are successful, so we haven’t started — as far as I know nobody has really started with any kind of a rational approach to designing drugs based on knowing the structure of the nuclear pore complex. But one can speculate that perhaps if one can interfere with import or export of certain protein molecules, in or out of the nucleus, you could achieve certain biomedical outcomes that are desirable.
And I’ll stick my neck out, I have to read more about the subject but I understand the Leptomycin B, one of the antibiotics that might be used in the near future, coincidentally does interfere with transport of p53 protein that’s involved in cell cycle regulation and whose malfunction may result in cancer.
There is a way to use Leptomycin B to keep more of the p53 inside the nucleus and therefore benefit from its regulative role to help treat cancer that way. So one can imagine more gains of this nature where either good players are camped inside the nucleus or more of them are pumped into the nucleus through judicious interference with the transport process made possible by really detailed understanding of how the transport through the NPC works.
Which in turn of course is made possible by knowing the structure of the NPC.
Miller: We’ve talked a lot about interactions so before I go I want to ask, is it possible to imagine being able to actually influence on protein, or a protein-protein interaction without creating some cascade effect with a lot of unintended consequences?
Sali: Well that’s certainly a big problem that we’re almost always faced with I suppose. In drug discovery you develop molecule hopefully that interacts or maybe usually inhibits a particular protein enzyme usually, but it almost never does. It has broader interactivity and then there are all sorts of side effects as a result.
So similarly, maybe more so, one would expect the specificity problems when you’re dealing with protein-protein interactions, and it’s just something that one has to be aware of and do the most to minimize or maximize — sometimes maybe you’d like to have a broad spectrum — a small molecule or a broad spectrum impact on protein-protein interactions, but it is an issue that’s combinatorial so-to-speak, in its nature and problematic and difficult to handle.
Miller: So What’s next for you now that this project is done, if in fact it is done?
Sali: Oh it’s not done (laughter). Always for one question answered there are ten more new questions. I think in this case we certainly want to get a high-resolution structure of the NPC; we want to develop data, use data that our friends can measure in the lab about the transport through the NPC and incorporate this data into, again, a quantitative model in a computer; we want to describe the evolution of the nuclear pore complex more accurately. We’ve done some work on that already with some intriguing findings but there’s more to do. There’s the question of how does the whole complex assemble 480 proteins, how do they come together when they first meet, so to speak, and then for us there’s always the need to generalize the computational method and apply it to as many other complexes, perhaps in collaboration with other biologists, as we can.
Miller: Well now I truly know what you’ll be doing everyday. Thank you for joining us on Science Café, congratulations on this work and good luck in the future.
Sali: Thank you very much.