A Speck of Spec: A Conversation with Al Burlingame

Time of flight. Liquid matrix secondary ion sputtering. Electron capture. Electrospray. Laser desorption. Beam transport.
Can anyone tell me what these have to do with CSI: Miami?
Nothing. And everything. You see, when those CSI lab sleuths snatch their paper readouts and name the distinctive elements in skin secretions or the pollen from an obscure plant — melodramatics aside — they are talking the language of mass spectrometry.
Mass spec, as it is commonly called, uses sensitive measuring techniques — based on electrical charge and mass — and some very expensive machines costing as much as $1 million each to identify and separate molecules, even if they exist in only infinitesimal amounts. Some popular uses have nabbed athletes on steroids or determined dioxin levels in contaminated fish.
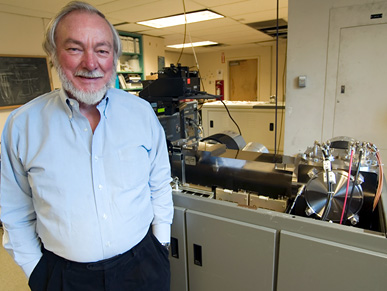
Mass spec can also reveal the structure and properties of molecules in complex biological samples, a particularly useful perspective when trying to understand how health and disease are influenced by the fluid and ceaseless activity inside a cell.
Not surprisingly for a technology so specialized, mass spec has its own language. Fortunately for many researchers at UCSF — and others around the world — that language has been spoken fluently for the last 30 years by Al Burlingame, PhD, director of the School of Pharmacy's Mass Spectrometry Facility: UCSF's own king of mass spec.
While the 70-year-old Burlingame resists being called UCSF's "king of spec," truth be told, his career has spanned several complete and total changes in technology, all of which have only whetted his curiosity. "We've gone from vaporizing samples to fast atom bombardment to electron capture," he explains. "And in doing so, sensitivity has increased by 10 to the third or more."
Sensitivity to what? you might ask. To the mechanics of life is one answer, which sounds nice, but doesn't explain much. How about this? We all know that proteins literally make us and also make us work. We also know that genes encode proteins and that genes are regulated or, in other words, that they are turned on and off. But as Burlingame reminds us, "There are processes by which proteins are switched on and off too by other proteins. These processes are not directly encoded in genomics. Defining those systems and processes are where we are focused."
Put another way, says Burlingame, "Knowing which genes are turned on and off in a cell does not reliably predict which proteins are there, how much of each protein is present, how the proteins have been chemically modified or how they might interact with other proteins." Proteins clearly affect how cells act, so knowing more about them is, well, key to knowing everything.
That brings us full circle to mass spec technology, which got a Nobel Prize-winning boost in the early 1990s when new techniques allowed bigger molecules, like proteins, to be more efficiently ionized, or electrically charged, and more specifically analyzed. "Years before, almost everything had to be vaporized first before you could do anything," says Burlingame. Now the sample can also be a solid or a liquid before it is injected into the vacuum chamber. In the case of protein samples, Burlingame explains, fragments are separated out beforehand by another technique called affinity chromatography. But that's another story.

What happens next is a powerful mix of physics and chemistry. But in essence, because the different molecules that make up the sample have different charges and mass, it is possible to analyze and identify them based on, for example, how long they stay airborne.

"We're basically profiling proteins," says Burlingame, "because techniques have advanced to the point where we can study them directly." And when you consider that humans likely host hundreds of thousands of proteins, one can imagine the embarrassment of riches spilling from the 10 machines in UCSF's Mass Spectrometry Facility, as well as from those in the 20 or so other similar facilities around the world.
It's a bioinformatics bottleneck, one reason why School of Pharmacy faculty have been hosting the software research engine, ProteinProspector, for nearly 10 years. The ProteinProspector, visited by 90,000 users each month, allows researchers to process very large data sets and to measure and compare several thousand proteins in a single experiment.

Burlingame and his colleagues are heavy users themselves. "We're trying to sort out this mess," he laughs, knowing that evolution has not made it easy. "Human cells are very difficult to deal with analytically, which is why we are trying to define what's happening by dividing and conquering their parts."
In the meantime, Burlingame has teamed up with scientists at UCSF and elsewhere to study everything from the p53 pathway in cancer to learning and memory. "Learning and memory is really an emerging field for us," Burlingame explains. Then, warming to the task, he adds, "There are at least 1,300 phosphorylation sites in the postsynaptic parts of brain regions associated with it. It's going to be quite a job."
But then, that's why mass spec has such a big name. When you set out to "globally penetrate the composition of human cells," as Burlingame reminds us, you had better be ready to think large.

Photos/Majed
Alternative content
Related Links
- UCSF Mass Spectrometry Facility
- UCSF ProteinProspector
- What is mass spectrometry?
- American Society for Mass Spectrometry
- Mass spectrometry
- Wikipedia
- Mass spectrometry
- William Reusch, Michigan State University
- Time of flight/ TOF mass spectrometer tutorial
- Jordan TOF Products, Inc.
- Time-of-flight mass spectrometry (TOF-MS)
- On-line Educational Hypermedia
- The Nobel Prize in Chemistry 2002: Revolutionary analytical methods for biomolecules